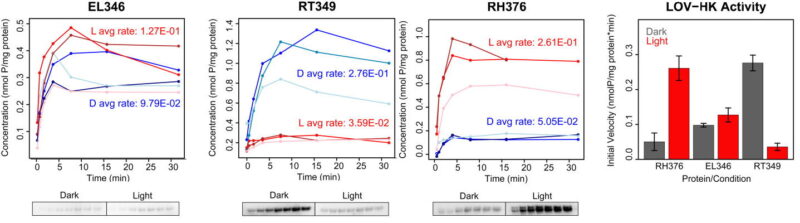
In our latest J. Biol. Chem. paper, we share some of our latest findings from a fantastic team led by Igor Dikiy and Danielle Swingle, who investigated how strongly homology across related proteins really translates into fundamental aspects like regulatory mode and quaternary structure. Starting from bioinformatics searches looking for proteins related to a monomeric light-activated histidine kinase (LOV-HK), EL346 (which we characterized here, here, and here after pioneering work by Trevor Swartz and Roberto Bogomolni), we thought we’d find a bunch of other light-activated kinases including some monomers.
We did … but not just that, either.
We found several light-activated LOV-HKs, but we also found a light-repressed LOV-HK too (just like engineered variants can, as Möglich and Moffat showed), showing that sequence conservation among these highly-related proteins doesn’t strictly retain the signaling logic. Quaternary structure is variable too, searching from our monomeric sequence found us dimers. Over in the related PAS-HKs – which are probably controlled by small molecule binding instead of light – we found a couple of proteins which interconvert between monomers and dimers, one of which is dimer-activated, the other dimer-repressed. Again, sequence to function conservation … yep, still there, but with some big caveats.
We’ve obviously got more work to do to sort out mechanism of some of these changes – the monomer/dimer aspects among the LOV-HKs are clearly not a simple change of one segment of the protein or the other – but we think the diversity of control & structure within a class of proteins is pretty cool and merited reporting now. Thanks to the editors and reviewers for JBC for helping us get the story out after a long journey to get here! KG