Gardner lab projects
The lab uses a combination of structural biology and biochemistry methods – from NMR / X-ray crystallography / cryo-EM to high-throughput screening to cellular assays – to probe the structure, function, and regulation of proteins involved in environmental sensing and signaling. Below, we list three general areas as examples of our approach; feel free to drop an email for more information –
Blue light photoreceptors
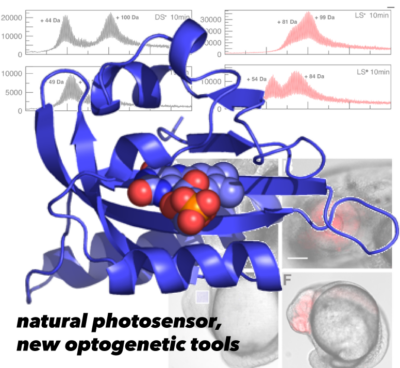 Nature utilizes photosensory proteins to allow cells to detect changes in light and react accordingly. We’ve focused our attention on two classes of such proteins which use flavin chromophores – FMN, FAD, or riboflavin – to detect changes in blue light, asking how photochemical changes at these cofactors are amplified by the surrounding proteins to achieve biochemical change. We’ve done most of this work in Light-Oxygen-Voltage (LOV) domains, starting from NMR studies of a plant phototropin LOV domain to determine how such changes control the structure of a part (“Jα helix”) that in turn controls enzymatic activity of this protein (Harper 2003, Harper 2004a, Harper 2004b). From this start, we’ve investigated similar changes in diverse full-length proteins – from bacterial histidine kinases (Correa 2013, Rivera-Cancel 2014, Dikiy 2019) and transcription factors (He 2002, Nash 2011, Zoltowski 2011, Rivera-Cancel & Motta-Mena 2012) to fungal controllers of G protein signaling (Glantz 2016, Glantz 2018) – to ask what elements are common among these proteins and which are different, and how these changes affect function in vitro and in living cells.
Nature utilizes photosensory proteins to allow cells to detect changes in light and react accordingly. We’ve focused our attention on two classes of such proteins which use flavin chromophores – FMN, FAD, or riboflavin – to detect changes in blue light, asking how photochemical changes at these cofactors are amplified by the surrounding proteins to achieve biochemical change. We’ve done most of this work in Light-Oxygen-Voltage (LOV) domains, starting from NMR studies of a plant phototropin LOV domain to determine how such changes control the structure of a part (“Jα helix”) that in turn controls enzymatic activity of this protein (Harper 2003, Harper 2004a, Harper 2004b). From this start, we’ve investigated similar changes in diverse full-length proteins – from bacterial histidine kinases (Correa 2013, Rivera-Cancel 2014, Dikiy 2019) and transcription factors (He 2002, Nash 2011, Zoltowski 2011, Rivera-Cancel & Motta-Mena 2012) to fungal controllers of G protein signaling (Glantz 2016, Glantz 2018) – to ask what elements are common among these proteins and which are different, and how these changes affect function in vitro and in living cells.
From this work, we’ve turned some natural photosensors into very useful ontogenetic tools, including one (EL222) that can turn genes on in a wide range of cell types using blue light (Motta-Mena 2014, Reade 2017, LaBelle 2021), leading to a spin-off company named Optologix. This work has also helped inspire a range of protein engineering applications in many labs where LOV domains are fused into proteins which aren’t normally light sensitive, turning them into new reagents for biotechnology (Losi 2018, and nicely surveyed by the hard working team at OptoBase). We’ve pursued a combination of approaches to make LOV domains better photosensors as part of this, too (Yao 2008, Strickland 2010, Song 2011, Freddolino 2013)
Complementing this work, we’ve also studied Blue Light sensors Utilizing Flavins (BLUF), which use similar chromophores and protein structures, but different photochemistry from the LOV domains. NMR and biochemical studies from our lab and others have shown similar structural changes as the LOV domains, albeit in ways that are different in some important ways (Jung 2005, Wu 2008, Wu 2009, Yuan 2011).
Sensing oxygen and nutrients
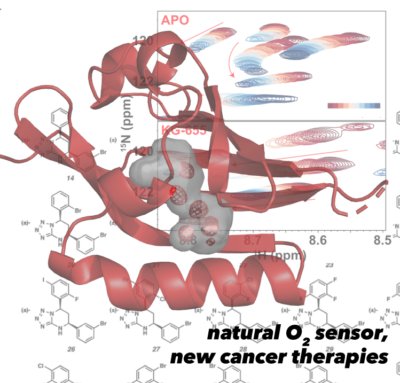 The LOV domains mentioned above turn out to be a subset of a much wider range of environmental sensory domains, the Per-ARNT-Sim (PAS) domains. We’ve investigated how these domains are used to control a number of human proteins with substantial biomedical impact.
The LOV domains mentioned above turn out to be a subset of a much wider range of environmental sensory domains, the Per-ARNT-Sim (PAS) domains. We’ve investigated how these domains are used to control a number of human proteins with substantial biomedical impact.
Foremost among this is our work on the Hypoxia Inducible Factors (HIFs), the focus of the 2019 Nobel Prize in Physiology to Kaelin, Semenza, and Radcliffe (Nobel Committee 2019). While normally controlled by an elegant combination of post-translational modifications, the loss of these controls leads to HIF overexpression that is correlated to cancer onset and progression. Using a combination of structural biology methods, we solved structures of parts of the human HIF-2 heterodimer that intriguingly included a large cavity within one of the HIF-2α PAS domains (Erbel 2003, Card 2005, Yang 2005, Scheuermann 2009, Key 2009, Correa 2016). Following parallels with the LOV domains mentioned above, we suspected that small molecules might bind and regulate HIF-2 function, which we demonstrated initially in vitro (Scheuermann 2009, Key 2009) and subsequently in cells together with Rick Bruick using small molecules found by high throughput screening (Scheuermann 2013, Rogers 2013, Scheuermann 2015). Our findings were key to the foundation of Peloton Therapeutics, Inc. and subsequent optimization of HIF-2α inhibitors for clinical trials as a route to treat certain kidney cancers (Chen 2016). This work continues at Merck, Inc. with the 2019 acquisition of Peloton Therapeutics. We continue to investigate HIF interactions other small molecule and protein partners, particularly with classes of transcriptional coactivator proteins (Partch 2009, Partch 2010, Partch 2011, Guo 2013, Guo 2015) that both give some insight into gene-specific HIF regulation and may guide additional therapeutic strategies.
In another disease setting, we have studied human PAS kinase – a serine/threonine kinase which is regulated by PAS domains – that controls nutrient flux with some interesting parallels to the plant phototropins mentioned above. Our work using solution NMR showed that one of the PAS domains from this protein binds both the kinase and artificial small compounds identified using NMR-based fragment screening, suggesting a way to artificially control this protein as a route for potential use in metabolic diseases (Rutter 2001, Amezcua 2002).
Switchable proteins and methods to study them
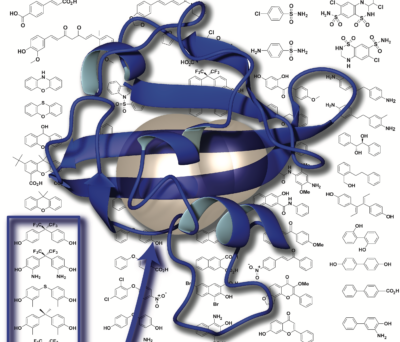 As part of our studies of natural and artificial control of protein structure, we’ve worked to establish good model systems for such work. We found one such system – the Y456T variant of the ARNT PAS-B domain – fortuitously as part of a mutagenesis project, finding that it exists in a slow 1:1 equilibrium between two conformations differing by an intriguing +3 slip in beta-strand register (Evans 2009a, Evans 2009b). We’ve recently demonstrated that we can control this equilibrium by binding small molecules that selectively bind only one of the two conformations (Xu 2021a) or by applying high pressure (Xu 2021b). Together this let us confirm some key mechanistic predictions – including that the protein must unfold to switch conformations – and lay the groundwork for artificial control. We’ve also extensively characterized one of the phototropin LOV domains (“AsLOV2″) as a robust model system for NMR and other studies of light-induced conformational changes (Harper 2003, Harper 2004a).
As part of our studies of natural and artificial control of protein structure, we’ve worked to establish good model systems for such work. We found one such system – the Y456T variant of the ARNT PAS-B domain – fortuitously as part of a mutagenesis project, finding that it exists in a slow 1:1 equilibrium between two conformations differing by an intriguing +3 slip in beta-strand register (Evans 2009a, Evans 2009b). We’ve recently demonstrated that we can control this equilibrium by binding small molecules that selectively bind only one of the two conformations (Xu 2021a) or by applying high pressure (Xu 2021b). Together this let us confirm some key mechanistic predictions – including that the protein must unfold to switch conformations – and lay the groundwork for artificial control. We’ve also extensively characterized one of the phototropin LOV domains (“AsLOV2″) as a robust model system for NMR and other studies of light-induced conformational changes (Harper 2003, Harper 2004a).
In parallel, we’ve developed and implemented a range of tools for studying the effects of physical and chemical perturbations on protein structure and dynamics, many of which use the unparalleled site-specific information provided by solution NMR. Most recently, this has included our use of high-pressure NMR methods to explore how conformational changes affect protein volume and compressibility (Xu 2021b), either under constant pressure or in a pressure jump mode. We’ve also used high pressure as a rapid way to screen proteins for internal cavities that might bind small molecule ligands (Gagné 2021). Our photoreceptor work has led us to develop methods to illuminate NMR samples in situ and monitor structural changes, either in steady state or in real-time kinetic modes with time-resolved NMR (Harper 2003, Harper 2004a). Finally, we’ve used NMR-based screening of a library of about 800 compound fragments to seed various drug discovery projects, including some of our HIF-2α and ARNT work mentioned above (Amezcua 2002, Best 2004, Scheuermann 2009, Key & Scheuermann 2009, Guo 2013, Xu 2021a).
